On February 3, a research paper entitled Resetting Histone Modifications During Human Prenatal Germline Development was published online in Cell Discovery (IF: 38). The research was done by Professor GAO Shaorong, Professor CHEN Jiayu, and Professor ZHANG Yong, from the School of Life Science and Technology of Tongji University jointly with Professor LI Xiaocui's team from the First Maternal and Infant Health Care Institute of Tongji University. This research revealed the epigenetic mechanism by which three core histone modifications (H3K4me3, H3K27me3, and H3K9me3) co-regulate gene and reverse transposon expression, X chromosome reactivation, and DNA demethylation escape events during the chromatin reprogramming of human primordial germ cells in a genomic hypomethylation state for months.
Mammalian primordial germ cells (PGCs) are precursors of germ cells that eventually develop into highly specialized gametes - sperm and eggs. The correct development of primordial germ cells is a necessary condition for maintaining genetic stability and continuity within a species, thereby ensuring that genetic and epigenetic information can be faithfully transmitted from one generation to the next. Primordial germ cells undergo very unique events in the process of development, including cell migration, reproductive ridge localization, gender determination, meiosis, and gamete maturation. More importantly, in this process, the transcriptome and an epigenetic group of primordial germ cells have undergone thorough and extensive reprogramming, making full preparations for subsequent gametogenesis and the establishment of cellular totipotency. However, constrained by the limited number of cells and molecular biological techniques, we have known little about the exact changes in the epigenetic group during the development of human PGC (hPGC).
In recent years, with the development of microomics technology and related bioinformatics, research teams both from home and abroad have conducted in-depth research on the dynamic changes in the transcriptome, DNA methylation group, and chromatin accessibility of hPGC. Studies have found that male and female hPGC exhibit gradual changes in gene expression during development, and exhibit heterogeneity in cell migration, mitosis, meiosis, and gametogenesis. At the same time, the chromatin accessibility and DNA methylation status in hPGC are similar to those of mice at corresponding developmental stages, indicating the conservatism of the two species in the dynamic changes in the reprogramming of primordial germ cells. However, current research on histone modifications in hPGC is mainly based on immunofluorescence staining, and its genome-wide distribution, dynamic changes, and correlation with other epigenetic modifications such as DNA methylation and chromatin accessibility are unclear. In addition, whether they are involved in transcriptional regulation and specific developmental events such as X chromosome reactivation and DNA demethylation escape during hPGC genome-wide hypomethylation remains to be addressed.
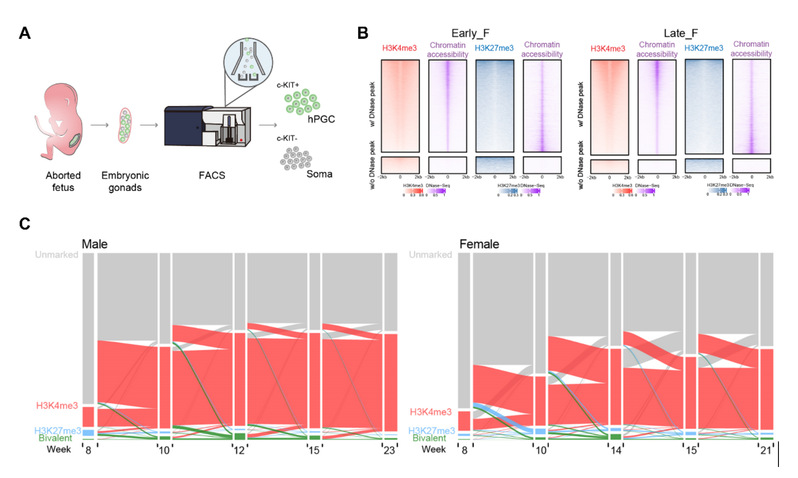
In this research, researchers used microcellular ULI-NChIP-seq technology to map for the first time a high-resolution genome-wide dynamic map of three key histone modifications (H3K4me3, H3K27me3, and H3K9me3) in male and female primordial germ cells during development from 7 to 23 weeks of gestation (Figure 1A). Studies have shown that H3K4me3 exhibits a typical promoter enrichment pattern. Unlike DNA methylation, H3K4me3 modification exhibits a high correlation with gene transcription and chromatin open state (FIGS. 1B and 1C). In contrast, H3K27me3 modification during the development of hPGC exhibits relatively low genome-wide enrichment, and it is mainly located in the H3K4me3 labeled region and negatively correlated with chromatin opening, playing an important role in regulating specific bivalent promoters related to development (FIGS. 1B and 1C). Interestingly, previous studies have shown that there is a unique X chromosome regulatory mechanism in the development of female embryos and germ cells, known as X chromosome inhibition (XCD). Among them, the X chromosome exhibits incomplete reactivation, which is regulated by lincRNA XACT rather than relying on the traditional lincRNA XIST. The research shows that there is no significant difference in the expression of XACT and XIST between men and women at 7-23 weeks of age in hPGC, and there is no significant difference in DNA methylation on the X chromosome between men and women, suggesting that neither of them can fully explain the regulation of X chromosome reactivation in female hPGC. Compared with the male X chromosome, the female X chromosome is highly enriched in H3K4me3 modifications, suggesting that reactivation of the X chromosome requires these active histone modification markers. Considering the overall hypomethylation status of hPGC in women and the enrichment of H3K4me3 signals on the X chromosome, why did the activation of two X chromosomes only result in a 1.6-fold increase in gene expression, rather than a 2-fold increase? Further research has found that female hPGC exhibits higher levels of H3K27me3 and H3K9me3 modifications on the X chromosome compared with male hPGC at the same developmental stage, suggesting that inhibitory histone modifications play an important role in limiting the complete reactivation of the X chromosome of female hPGC.
Dynamic changes of histone modifications in human primordial germ cells
It is worth noting that although hPGC exhibits DNA demethylation at the early stages of development (beginning at 4 weeks of gestation), there are certain specific regions that consistently exhibit high levels of methylation, known as demethylation escape regions. In this study, it was found that H3K9me3 was mainly labeled in these escape regions rather than in the regions where demethylation occurred. In addition, heterochromatin-modified H3K27me3 may play a synergistic role in consolidating methylation in these escape regions. At the early stages of development, co-markers of histones H3K9me3 and H3K27me3 can be detected simultaneously on these demethylated escape regions, indicating that these two core histone markers are sufficient to protect these escape regions from complete demethylation during development, while the entire genome is still undergoing a continuous and thorough demethylation process. In contrast, co-markers of H3K9me3 and H3K27me3 rarely occur simultaneously in the demethylated region, and the expression level is high. Given the global DNA demethylation and the synergistic activation of H3K4me3 on gene expression in primordial germ cells during development, this study revealed that inhibitory histone modifications H3K9me3 and H3K27me3 are jointly involved in the stable maintenance and transcriptional regulation of demethylated escape regions in hPGC. It is worth mentioning that recently, the Surani Research Group of the University of Cambridge, UK, published a research paper entitled Epigenetic resetting in the human genome line entails histone modification remodeling in Science Advances, which also shows that inhibitory histone modifications H3K9me3 and H3K27me3 have important regulatory effects on hPGC retrotransposon elements in the whole genome hypomethylation state.
Professor CHEN Jiayu, Professor ZHANG Yong, and Professor GAO Shaorong of Tongji University are the co-authors of this paper while Assistant Professor GAO Rui, Ph.D. student ZENG Shiyang, and Professor YANG Dongxu and LI Xiaocui are the co-first authors. Professor LIU Wenqiang, Professor GAO Yawei, Dr. BAI Dandan, Dr. Zhang Linfeng, Dr. CHEN Chuan, attending physician HONG Wei, and Deputy Chief Physician WANG Beiying have made essential contributions to this study. This work has received important support from projects such as the National Natural Science Foundation of China and the National Key Research and Development Plan.